sbml-diff
sbml-diff is a tool for visually representing a single SBML model, or for visually comparing 2 or more SBML models. It can be used in three ways:- As a command-line tool
- As a python package
- through our web interface (example comparison between two or three models.)
@article{ScottBrown2016,
doi = {10.1021/acssynbio.6b00273},
url = {https://doi.org/10.1021%2Facssynbio.6b00273},
year = {2016},
month = {dec},
publisher = {American Chemical Society ({ACS})},
author = {James Scott-Brown and Antonis Papachristodoulou},
title = {sbml-diff: A tool for visually comparing {SBML} models in synthetic biology},
journal = {{ACS} Synthetic Biology}
}
Resulting images
Many options are provided to adjust the resulting images, but there are 3 major types of representation.
Features are associated with models using color: if a feature is present in a single model it is assigned that model's color, and if it is present in some (but not all) models it is drawn in black. Features present in all models it is drawn in grey, so that it visually fades into the background.
Default rendering
In this view, each element of an SBML model (species, reactions, rules, events) are represented by a node in a node-link network representation.
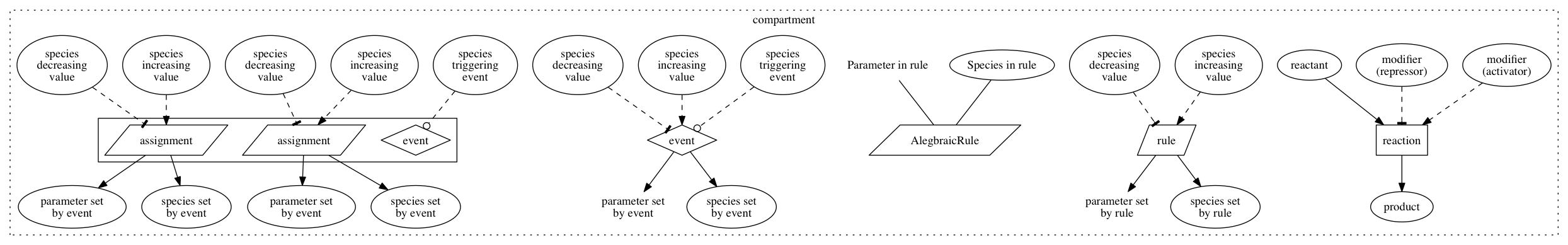
Reactions or rules that appear in more than one model, but with different kinetic laws, are displayed with a grey background. Colouring is used to indicate whether each node and edge is common to all models (grey), some but not all models (black), or a single model (a colour specific to that model).
The user can choose between labelling reaction nodes with the corresponding reaction name, kinetic law, or name and rate law, or leaving them unlabelled.
Cartoon rendering
This rendering differs from the default rendering in two main ways. Transcription reactions are represented by SBOLv glyphs (rather than the generic rectangular reaction node). If a transcription reaction is followed by a translation reaction, and the intermediate species is not involved in any other reactions (except degredation), the transcription-translation is represented as a single reaction.
Abstract rendering
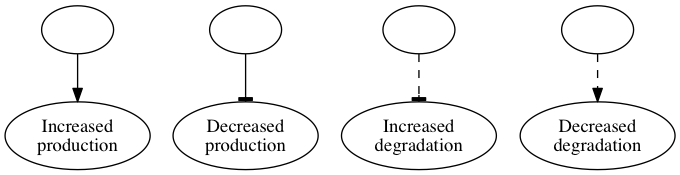
In this rendering, all nodes represent species. Arrows indicate that one species increase the production, decreases the production, increases the degredation, or decreases the degredation of another species.
Limitations
Reactions and species are compared based on their ID. Additionally, two elements are treated as being the same if both have exactly the same (non-empty) set of MIRIAM identifiers.
The arrowheads indicating activation/repression of a reaction by a ModifierSpecies may be misleading if the corresponding kineticLaw is not monotonic, or if the kineticLAw could represent either activation and repression, depending on the exact parameter values chosen.